Projects
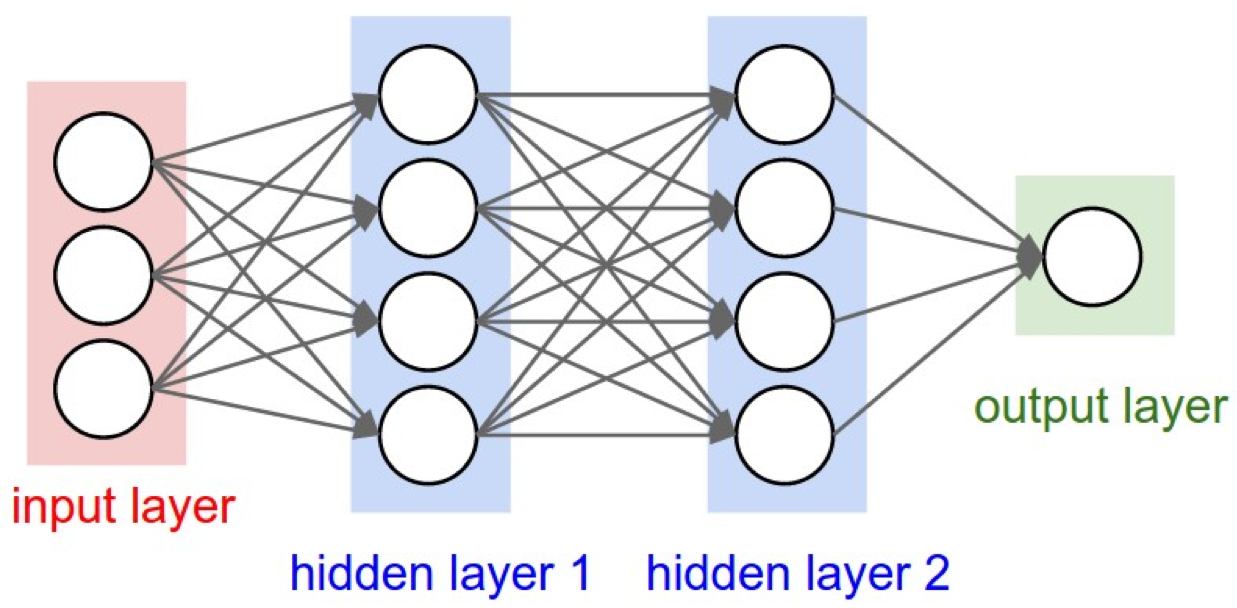
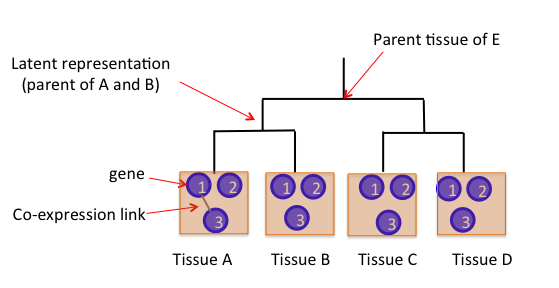
Interpreting deep learning models to understand gene regulation and cellular differentiation
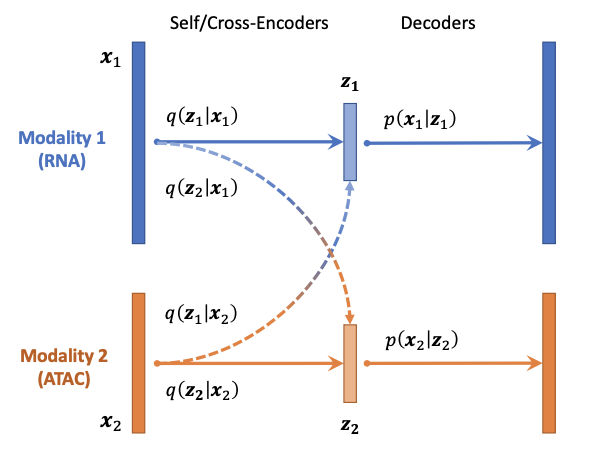
Models for single cell genomics
Relevant publications include CLUE.
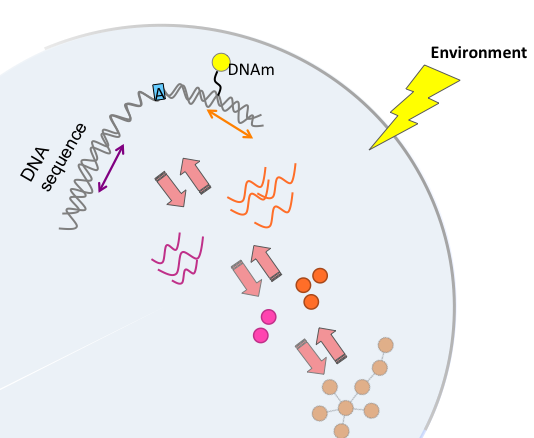
Understandding genetic variation and its contribution to human disease
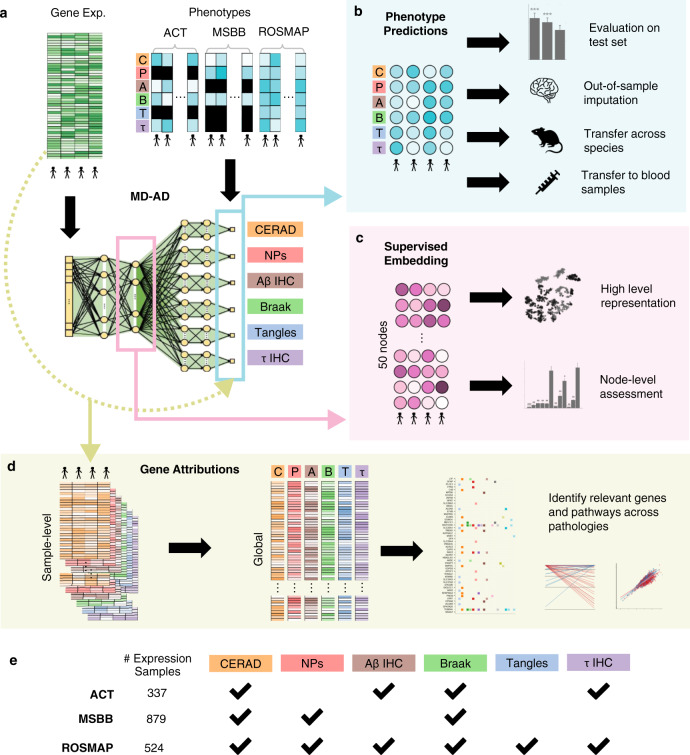
Multi omics analysis of complex disease